Nucleic acid extraction is the first step in pathogen nucleic acid testing and aims to isolate nucleic acids from biological samples and remove substances such as proteins, sugars, and lipids that may interfere with subsequent analyses to ensure the integrity of the nucleic acids.
Traditional nucleic acid extraction methods include phenol-chloroform, boiling lysis, centrifugal columns and magnetic beads.
The phenol-chloroform method treats the sample with phenol-chloroform, which causes nucleic acids to dissolve in the aqueous phase, while polysaccharides and lipids dissolve in the organic phase and proteins precipitate at the interface of the two phases. Subsequently, nucleic acids are precipitated using ethanol and separated by centrifugation. Although this method yields high purity nucleic acids, it is complex and prone to environmental contamination.
Boiling lysis extracts nucleic acids by heating, boiling and high-speed centrifugation, but may result in loss of DNA or incomplete lysis, and may also trigger cross-contamination, affecting assay accuracy.
The centrifugal column method utilizes a silica matrix to adsorb specific DNA molecules, while RNA and proteins are passed through the column and treated with different conditions of salt and pH to ultimately purify the nucleic acids. The method is simple and easy to use, but not suitable for high throughput and automation.
The magnetic bead method uses surface-modified superparamagnetic nanoparticles that bind specifically to nucleic acids, and the extraction process eliminates the need for centrifugation and complex reagents, making it suitable for automated extraction, despite its high cost.
In addition to these common methods, sonication, repeated freeze-thawing, enzymatic digestion and hypotonic lysis are also effective in extracting nucleic acids, but they usually require longer time, high cost and complicated operation, and are not suitable for use under limited conditions.
Microfluidic Chip Nucleic Acid Extraction utilizes microfluidic technology to achieve rapid isolation and purification of nucleic acids by means of microdroplets of sample and reagents in a fluidic channel. This method is simple and fast, making it ideal for on-site high-throughput testing.
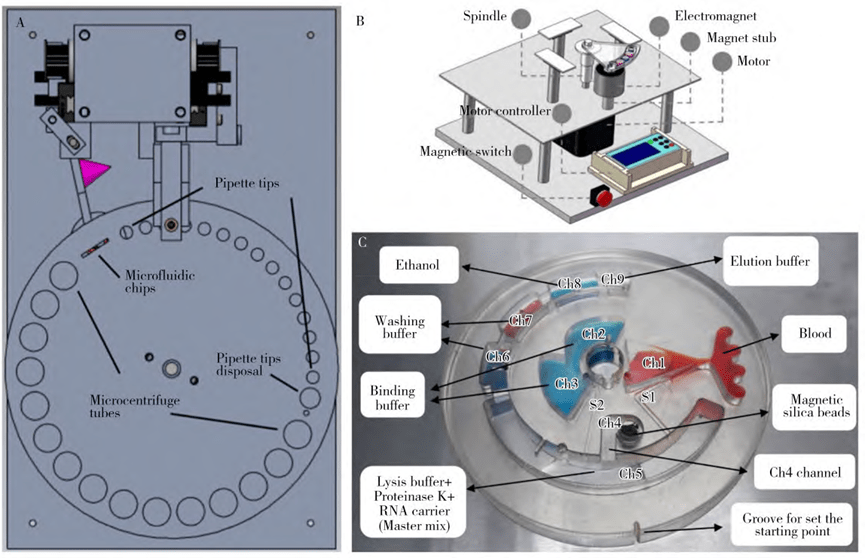
1. Magnetic bead-based microfluidic nucleic acid extraction
The magnetic bead method is a high-purity nucleic acid extraction method that has emerged since the 1980s and is now widely used.
Magnetic beads consist of metal particles (e.g., Fe2O3, Fe3O4), outer polymer materials (e.g., polystyrene, polyvinyl chloride), and functional groups (e.g., -NH2, -COOH, -OH, -CHO).
Nucleic acids are released from the sample in the presence of the lysate, which then binds to the beads to form “nucleic acid-bead complexes” with the help of electrostatic, hydrophobic and hydrogen bonding forces.
Under the action of an applied magnetic field, the complexes are separated, cleaned and eluted to finally obtain the desired nucleic acids.
The extraction efficiency is affected by a number of factors, such as bead incubation time, magnetic field strength, and ethanol residue.
Compared with the traditional magnetic bead extraction method, the microfluidic chip-based magnetic bead extraction method is improved in terms of automation and extraction efficiency.
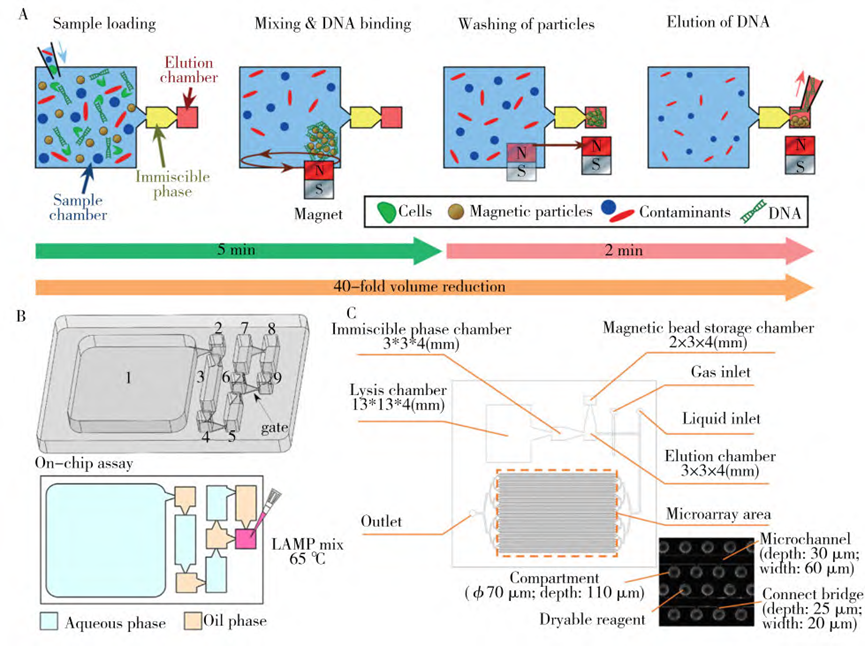
2. Microfluidic nucleic acid extraction based on surface tension assisted immiscible filtration method
Surface tension-assisted immiscible phase filtration (IFAST) technology enriches, washes, and elutes nucleic acids in just a few minutes by placing multiple aqueous-oil barriers (“gated” intervals) within the microfluidic chip and dragging the beads across these barriers using a handheld magnet.
The IFAST-based nucleic acid extraction method has the advantages of low cost, simplicity and rapid reaction without complex washing steps, thus reducing the risk of contamination and nucleic acid loss.
The method requires pre-loading of reagents and samples on the chip at the time of use, and the transfer of the beads is accomplished by manually controlling the magnet.
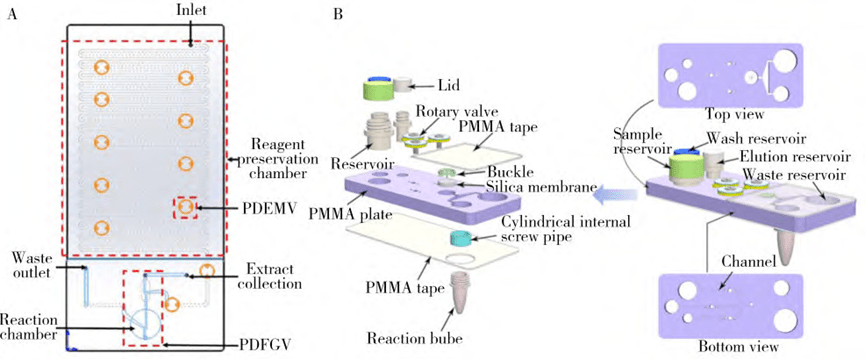
3. Silicon matrix-based microfluidic nucleic acid extraction
Silicon-based materials exist mainly in the form of glass powders, microfibers, microcolumns, and thin films. Silicon-based microfluidic chips have excellent thermal stability, good thermal conductivity, and strong resistance to organic solvents.
However, silicon materials are brittle, expensive, poorly electrically insulating, and have more complex surface chemistry, so problems such as excessive hardness, processing difficulties, and nonspecific adsorption need to be addressed in microfluidic chip applications.
The figure below illustrates a serpentine tubular microfluidic chip that utilizes ultrasonic lysis and is based on a silica membrane for nucleic acid extraction.
The reagents are pre-encapsulated in a serpentine tube and the automatic release of the reagents is achieved through a membrane valve, enabling the extraction of Hepatitis B Virus (HBV) and Human Immunodeficiency Virus (HIV) viral nucleic acids from serum samples.
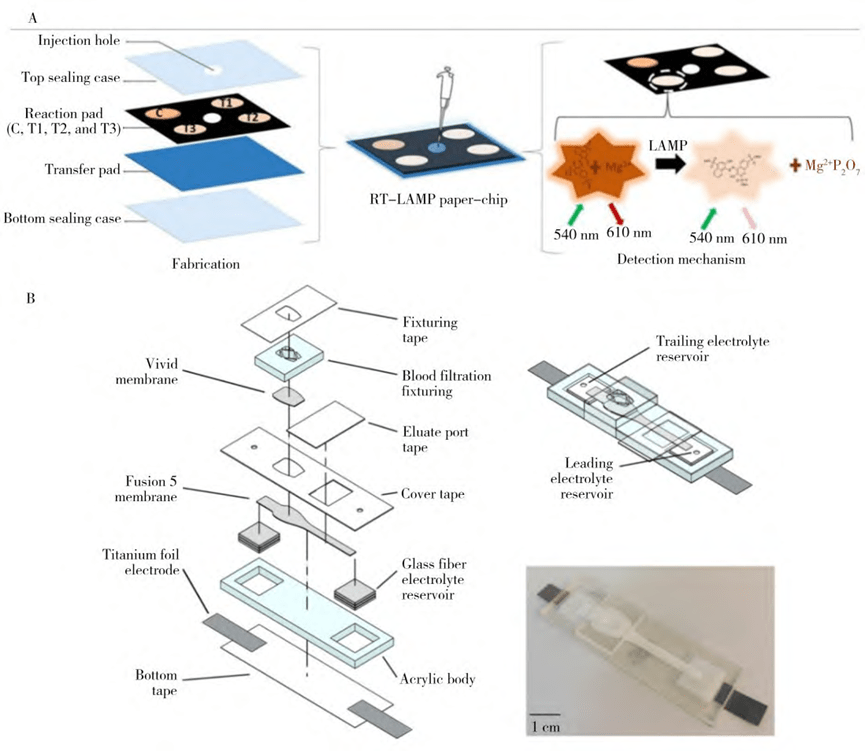
4. Paper-based microfluidic nucleic acid extraction
The paper-based method of extracting nucleic acids is based on the principle that DNA molecules are negatively charged and can be quickly adsorbed onto the surface of paper fibers in an alkaline salt solution.
Through a simple washing process, the DNA is retained on the surface of the paper fiber and eventually released into the amplification reaction solution for subsequent analysis.
Paper-based microfluidic chips use filter paper as a substrate in place of silicon, glass, or polymer materials and utilize techniques such as photolithography, wax printing, and inkjet printing to create microfluidic channels with alternating hydrophilic and hydrophobic structures.
Certain research teams have developed a paper-based microfluidic device capable of extracting RNA from serum samples for Zika, dengue and chikungunya viruses.
The extracted nucleic acids can be amplified on a paper chip by reverse transcription loop-mediated isothermal amplification, enabling simultaneous extraction and detection of viral RNA.
Paper-based microfluidic chip has the advantages of low cost, no external drive device, fast extraction speed, low reagent consumption and low environmental pollution.
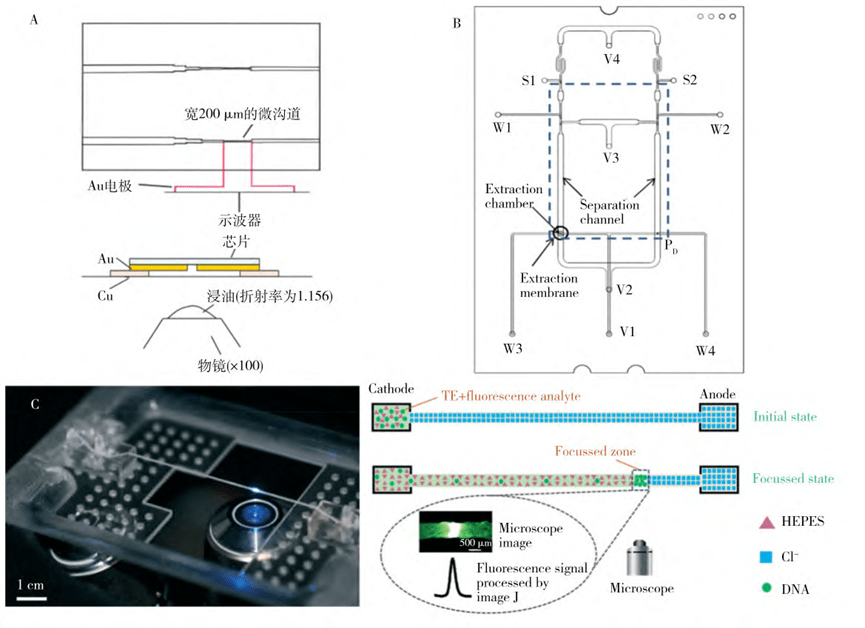
5. Electrophoresis-based microfluidic nucleic acid extraction
Nucleic acid electrophoresis is usually performed in agarose gels or polyacrylamide gels.
By adjusting the concentration of the gel, the pore size of its molecular sieve mesh can be changed, which in turn affects the separation of nucleic acids.
Under the action of electric field, due to the negative charge of nucleic acid, nucleic acid molecules will migrate to the anode, so as to realize the separation and purification of nucleic acid, which is especially suitable for the separation of nucleic acid fragments of different lengths.
In recent years, isokinetic electrophoresis has been widely used in the selective separation and enrichment of nucleic acids.
This approach creates discontinuous electrolyte systems by using electrolyte ions that move at different speeds, including slower terminating electrolyte ions and faster leading electrolyte ions, and modulates the migration of the target molecules by an electric field.
It was shown that a disposable integrated microfluidic device, combining isokinetic and gel electrophoresis, is capable of preconcentrating and separating multiple DNA fragments and extracting pure DNA fragments during on-chip electrophoresis.
About Us
DINGXU (SUZHOU) MICROCONTROL TECHNOLOGY CO., LTD. is a high-tech enterprise in the field of microfluidics, specializing in the development, production and sales of microfluidic control solutions. Our core business covers microfluidic chip customization, surface modification technology services, microfluidic supporting instruments and microfluidic chip processing equipment development and production. We are committed to providing reliable microfluidic solutions to our global customers through technological innovation and high quality products to help them realize more accurate, reliable and efficient experiments and applications in the fields of biology, medicine, life sciences and environmental monitoring.
© 2025. All Rights Reserved. 苏ICP备2022036544号-1













