1. Introduction to Nucleic Acid Amplification Technology
Nucleic acid amplification technology is an experimental method for nucleic acids (both DNA and RNA) designed to amplify a large number of target nucleic acids from a small sample of starting nucleic acids.
Among these, the polymerase chain reaction (PCR) was the first and remains the most popular amplification technique used to amplify and detect low abundance nucleic acids.
Isothermal cycling amplification eliminates the need for thermal cycling of PCR and allows for fast and efficient amplification at a constant temperature, making isothermal cycling amplification a favorable replacement for PCR.
Isothermal nucleic acid amplification techniques usually include enzyme-assisted amplification and enzyme-free amplification.
Enzyme-assisted amplification strategies include rolled loop amplification (RCA), loop-mediated isothermal amplification (LAMP), and exponential amplification reaction (EXPAR); enzyme-free amplification methods include strand displacement amplification (SDA), hybridization chain reaction (HCR), DNAzyme, entropy-driven circuits (EDC), and catalytic hairpin self-assembly (CHA).
Meanwhile, isothermal amplification technology detects a wide range of targets, including proteins, cells, small molecules and ions.
2. Isothermal enzyme-assisted signal amplification techniques
1) Rolling Circle Amplification (RCA)
Rolling Circle Amplification (RCA) is a highly specific and sensitive nucleic acid amplification technique that utilizes a circular DNA template and DNA polymerase for continuous amplification.
The basic principle is that by adding primers to a specific starting point of a circular DNA template, DNA polymerase initiates amplification and synthesizes a new DNA strand by rolling it along the template.
This process is repeated over and over, generating a large number of long strands of repetitive single-stranded DNA products.
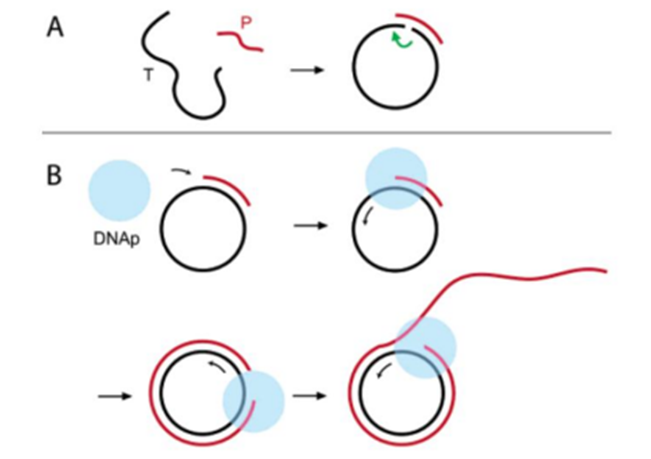
Unlike conventional PCR, RCA amplification does not require a thermal cycling process and thus can be performed at room temperature, making it easy to operate and the large amount of amplified product makes it suitable for the detection of low concentrations of target nucleic acids.
RCA not only has high sensitivity, but also can be targeted by designing specific probes, so it has a wide range of applications in the fields of molecular diagnosis, pathogen detection, genetic analysis and biosensor development.
The long chain structure of RCA amplification products can also be used in microfluidic devices, nanotechnology and single molecule assays.
2) Loop-mediated isothermal amplification (LAMP)
Loop-mediated isothermal amplification (LAMP) is a highly sensitive isothermal nucleic acid amplification technique that employs specific primers and polymerases to amplify DNA at a constant temperature.
LAMP amplifies target DNA sequences rapidly and efficiently by binding four or six specific primers to the DNA template to form a complex secondary structure with an inner and outer ring structure.
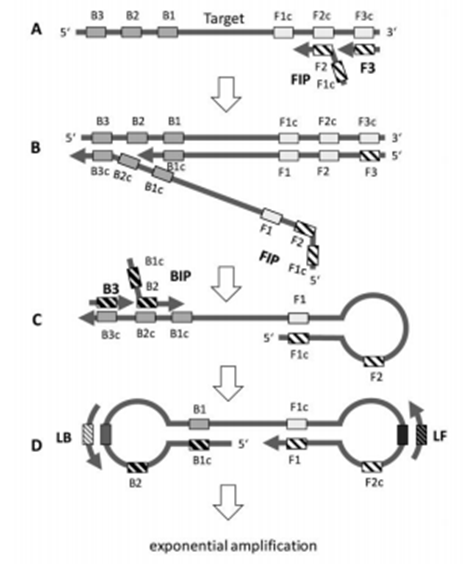
The core advantage of this method is the isothermal reaction, which does not require complex temperature cycling equipment and can be operated at ambient or lower temperatures.
The key to the LAMP amplification reaction is the formation of cyclic amplification products, which are generated by the synthetic activity of the polymerase in a continuous cyclic process that produces a large number of DNA products, usually in a characteristic “dendritic” structure.
LAMP technology has very high amplification efficiency and sensitivity, detecting very low concentrations of target DNA in a short period of time, and the amplification products can be visualized by the naked eye, e.g., by using changes in the turbidity of the solution or by dye fluorescence.
3) Exponential Amplification Reaction (EXPAR)
The Exponential Amplification Reaction (EXPAR) is an isothermal nucleic acid amplification technique that utilizes short primers and specific enzymes for rapid amplification at a constant temperature.
The basic principle of EXPAR is to cause the target DNA or RNA sequence to grow exponentially in a short period of time by triggering a series of specific amplification reactions.
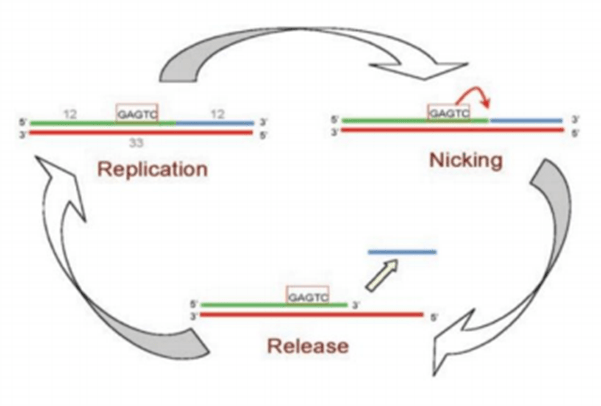
The key to the method is two primers: one primer binds to the target DNA template and initiates the amplification reaction, while the other primer binds to the resulting amplification product and facilitates the synthesis of a new strand through a series of catalytic reactions.
As the reaction proceeds, the product is amplified exponentially, eventually generating a large number of target sequences.
EXPAR is characterized by its rapidity and efficiency, and usually requires only a short time (e.g., within 30 minutes) to complete the amplification.
Since no thermal cycling equipment is required, EXPAR is also suitable for on-site applications in resource-limited environments.
3. Isothermal enzyme-free driven signal amplification technology
1) Entropy Driven Circuit
Entropy-driven circuits (EDC) are self-organizing systems based on the principle of entropy change for simulating and implementing complex biomolecular reactions or information processing processes.
The design of such loops is inspired by the principle of entropy increase in thermodynamics, which utilizes entropy changes between molecules to drive self-regulation and information transfer in a system.
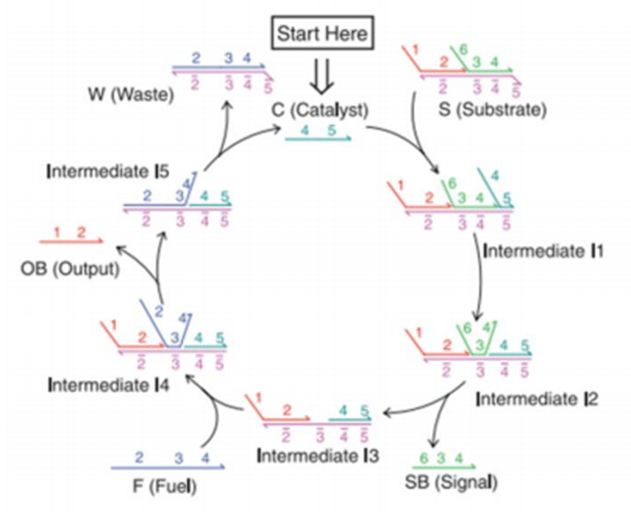
In entropy-driven circuits, molecules in a system (usually biomolecules such as DNA, RNA, or proteins) reduce entropy in localized regions through specific interactions and structural changes, while leading to an increase in the overall entropy of the system.
This entropy change drives reactions or information flow to accomplish complex functions such as self-replication, signal transduction, and computational processing. Entropy changes not only play a role in physical and chemical reactions, but also encode and transmit information at the molecular level.
2) Hybridization Chain Reaction (HCR)
Hybridization Chain Reaction (HCR) is a technique for signal amplification by specific hybridization reactions between nucleic acid molecules, mainly used to detect target nucleic acid sequences with high sensitivity.
The basic principle is to utilize two specifically designed primers that have complementary sequences that can interact in the presence of the target nucleic acid to form an autocatalytic cascade reaction that results in signal amplification.

Unlike traditional amplification techniques such as PCR, HCR does not rely on the involvement of enzymes and therefore avoids the temperature sensitivity of enzymes and is easier to operate.
The significant advantage is the extremely high sensitivity and specificity, as the reaction is carried out by hybridization and cascade reaction of specific primers, which can effectively amplify weak signals.
In addition, the products of HCR reactions are usually long-chain structures that can be detected by changes in solution turbidity, fluorescent labeling, or other methods.
3) Catalyzed Hairpin Self-Assembly (CHA)
Catalytic hairpin assembly (CHA) is a self-assembly amplification technique based on the structure of nucleic acids and catalytic reactions that is widely used for molecular detection and analysis.
The basic principle is to utilize a specifically designed DNA hairpin structure that triggers an autocatalytic reaction in the presence of the target nucleic acid, leading to an amplification reaction.
CHA reactions typically involve two hairpin DNA probes (called hairpin primers) that maintain a stable hairpin structure and do not bind to each other in the absence of a target.
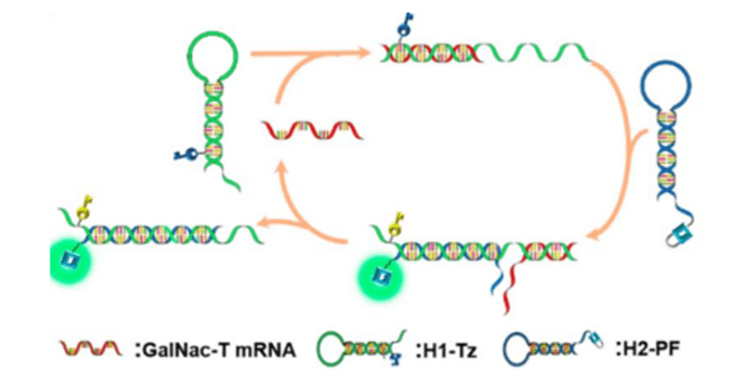
When the target DNA or RNA sequence is present, it binds to the two hairpin primers, prompting them to unfold and bind to each other to form a new complex.
This process activates a cascade of autocatalytic reactions, allowing more hairpin primers to be involved in the reaction, gradually forming a large number of double-stranded products, leading to amplification of the target sequence.
CHA is characterized by a reaction that does not require enzyme catalysis and relies on cascade amplification by hybridization and assembly reactions between nucleic acid molecules.
4) DNAzyme
DNAzyme is a class of catalysts consisting of DNA molecules with the ability to catalyze specific biochemical reactions, similar to protein enzymes.
Unlike conventional enzymes, DNAzyme consists of artificially designed DNA sequences that form specific catalytic activity centers through appropriate folding and structure, enabling them to perform catalytic functions similar to those of protein enzymes.
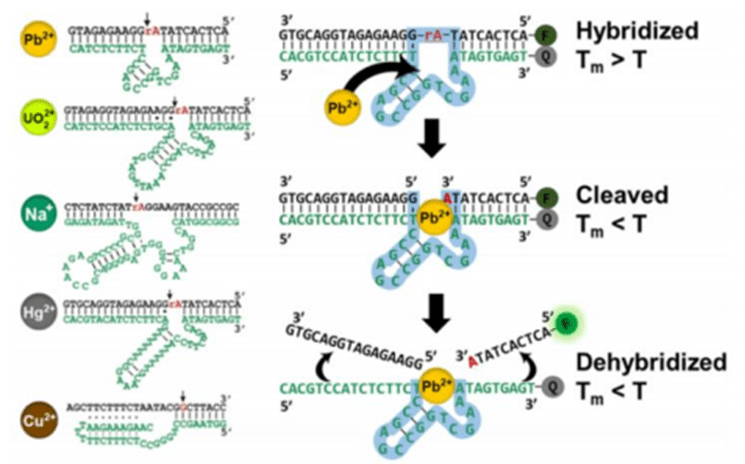
The function of DNAzyme is usually dependent on the folding of its three-dimensional structure, which can form a catalytic site through a specific arrangement of nucleic acid sequences, thus facilitating the conversion of substrates.
DNAzyme can catalyze a variety of chemical reactions, including nucleic acid hydrolysis, phosphorylation, dephosphorylation, and metal ion catalysis.
One of the most famous DNAzyme is the “10-23 DNAzyme”, which catalyzes the hydrolysis of RNA, a discovery that has provided an important tool in the fields of gene targeting and RNA interference.
© 2025. All Rights Reserved. 苏ICP备2022036544号-1













