PCR technology
Polymerase chain reaction (PCR) is a technique to amplify target nucleic acids in vitro, which has been widely used in genetic engineering, medical molecular diagnosis, environmental testing and other fields.
PCR technology utilizes DNA polymerase to amplify the initial target nucleic acid millions to billions of times under appropriate conditions through a thermal cycling process of denaturation, annealing and extension.
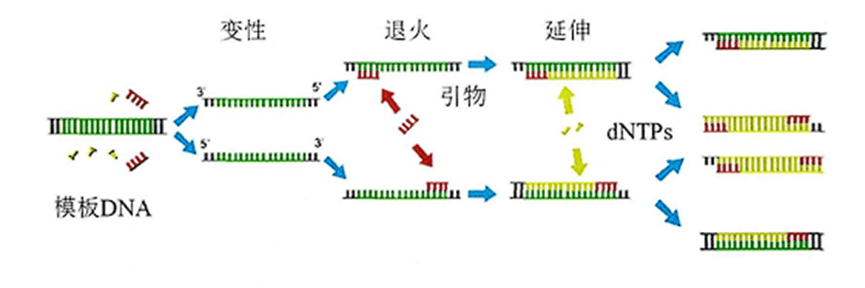
Based on their quantitative performance, they can be simply classified into three categories: qualitative PCR, real-time fluorescence quantitative PCR (qPCR) and digital PCR (dPCR).
1. Principles and features of dPCR technology
The dPCR technique consists of three main components: the DNA amplification reaction, fluorescence signal detection and data analysis.
Prior to the DNA amplification reaction, dPCR dilutes the template to the level of individual DNA molecules by limit dilution and assigns each DNA molecule to a different reaction unit for PCR amplification.
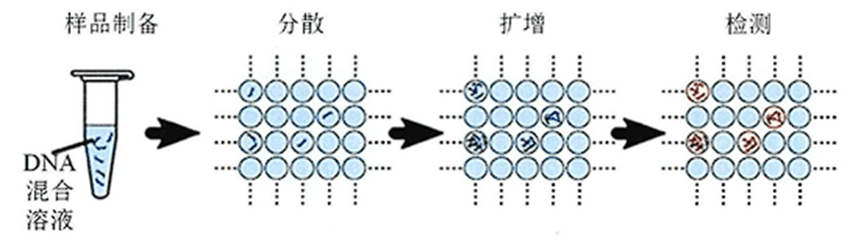
After the amplification is completed, the fluorescence signal of each reaction unit is collected. If the reaction unit has a fluorescence signal, it is recorded as 1; if there is no signal, it is 0.
The reaction unit containing the fluorescent signal contains at least one target DNA molecule, but may have multiple molecules.
Therefore, it is necessary to apply a Poisson distribution calculation that combines the number of units positive for fluorescence signal, the number of total reaction units and the template dilution to finally obtain the copy number of the target DNA molecule.
The key to dPCR is how each target DNA molecule is independently assigned to a different reaction unit.
Depending on the implementation, dPCR can be divided into two types: microdroplet digital PCR (ddPCR) and microarray chip digital PCR (cdPCR).
1) Microtitre Digital PCR
Microdroplet digital PCR utilizes the principle that water and oil are immiscible, using oil as the continuous phase and water as the dispersed phase, and generating tiny droplets by a specific method, these droplets become independent reaction units.
The microdroplet generation device enables the generation of tens of thousands of nanoliter- or picoliter-scale droplets, each encapsulating a single DNA molecule and PCR reagent.
DNA amplification was carried out independently in each droplet, and after amplification, the fluorescence signal of each droplet was detected and analyzed.
2) Microarray chip digital PCR
Microarray-on-a-chip digital PCR works by designing a chip that independently distributes gene amplification systems at the nano-level and below into a pool of microchannels, followed by DNA amplification and fluorescence signal detection.
According to the way the chip is designed, microarray PCR chips can be categorized into three types: integrated micropump valve chip, array micropool chip and slide chip:
1. Integrated micropump-valve chip: design liquid and gas channels on the chip through multilayer soft etching technology, use micropump valve to control the flow of liquid and distribute PCR amplification system to independent reaction units.
2. Array microcell chip: multiple microcells are etched on the surface of the chip, and templates and amplification reagents are precisely distributed to each microcell to form a large number of independent gene amplification reaction units.
3. Sliding chip: Microfluidic channels and reaction units are designed on a glass chip, and the PCR system is introduced from the liquid channels to each reaction unit by sliding the chip.
3) Characteristics of dPCR technology
Compared with first-generation PCR and second-generation qPCR techniques, dPCR technology has significant advantages, such as high sensitivity, high specificity and high accuracy:
1. High sensitivity: dPCR can detect individual nucleic acid molecules with a detection limit as low as 0.001%.
2. High specificity, accuracy and precision: dPCR can effectively differentiate samples with small differences in concentration and ensure accuracy.
3. Wide range of detection: Since the samples are diluted to the limit, they are effectively resistant to interference from inhibitors such as salts and preservatives, making them suitable for the detection of complex samples such as food, animal blood and plant tissues.
4. Absolute quantification: dPCR can directly quantify the target DNA molecules without the need for internal reference genes or standard curves.
5. Flexible reaction volume: the reaction volume can be adjusted from a few microliters to tens of microliters, which is suitable for quantitative detection of different samples.
2. Application of dPCR technology in food safety nucleic acid detection
1) Detection of Foodborne Pathogenic Microorganisms
Bacterial Detection: For example, E. coli, Salmonella, Listeria, Campylobacter, etc. dPCR can detect pathogenic bacteria in food even in low numbers by targeting specific genes (e.g., rRNA genes or virulence factor genes) for quantitative analysis. For pathogenic microorganisms in food, dPCR can provide more precise quantitative results to help warn of potential health risks.
Virus detection: such as Norovirus, Hepatitis A virus, Gastroenteritis virus, etc. These viruses are often transmitted through food, especially seafood and raw food. dPCR can accurately detect the genetic material of viruses (RNA/DNA) in food samples, and with high sensitivity, it can recognize viruses with very low copy number, guaranteeing rapid detection and control of viruses in food.
2) Plant-derived food ingredient testing
Detection of genetically modified ingredients: With the widespread cultivation of genetically modified crops, the presence of genetically modified ingredients in food has become an important issue for food safety. dPCR can detect the presence and content of genetically modified ingredients by targeting specific genetically modified DNA fragments (e.g. GMO marker genes). Compared with traditional PCR, dPCR does not need to rely on standard curves and provides higher quantitative accuracy, which is especially effective for low levels of GMO ingredients.
Pesticide Residue Gene Detection: dPCR can detect gene markers for pesticide residues or their degradation products in plant-derived foods, avoiding the tedious and time-consuming routine chemical analysis.
Food ingredient labeling: For example, testing food products for allergenic ingredients (e.g., soy, peanuts) to ensure consumer health and safety.
3) Detection of food ingredients of animal origin
Adulteration and species identification: dPCR can be used to characterize the authenticity of meat, such as identifying beef, lamb, pork, etc. for adulteration, or even detecting species-specific components in meat products. This is important for meat adulteration that often occurs in the market.
Detection of pathogens of animal origin: For pathogens in food of animal origin (e.g., mad cow disease, swine fever, etc.), dPCR allows for rapid and precise quantification of viruses or bacteria. For example, detecting the presence of swine fever virus in pork or avian influenza virus in poultry meat.
Antibiotic and drug residues: Antibiotic and hormone drug residues are a major concern for animal-derived food safety. dPCR can indirectly or directly indicate drug residues in food by detecting the expression of specific genes, which provides a more reliable basis for food regulation.
4) Genetically Modified Food Ingredient Testing
Quantification of GM ingredients: dPCR can be used to detect DNA markers in GM crops and accurately quantify the proportion of GM ingredients. For example, it can be used to detect the amount of GM crops such as corn, soybeans, cotton, etc. in food products to ensure that they do not exceed regulatory safety limits.
Detection of low concentration of genetically modified (GM) ingredients: While traditional PCR may give false negative results at low copy number, dPCR is able to directly calculate the gene copy number, and can accurately detect GM ingredients even if the content of GM ingredients is very low (e.g., less than 0.1%) in the food.
5) Food Virus Testing
Virus detection and quantification: dPCR can be used to detect viral DNA or RNA in food with high sensitivity and specificity, enabling detection at low viral load. For the monitoring of norovirus, hepatitis A virus, etc. in food, dPCR provides an efficient and accurate method.
Safety assessment: By quantitatively analyzing the amount of viruses in food, dPCR can help assess the risk of food-borne viruses, especially in the event of epidemics, enabling timely identification of the source of contaminated food and reducing the risk of food-borne viruses.
About Us
DINGXU (SUZHOU) MICROCONTROL TECHNOLOGY CO., LTD. is a high-tech enterprise in the field of microfluidics, specializing in the development, production and sales of microfluidic control solutions. Our core business covers microfluidic chip customization, surface modification technology services, microfluidic supporting instruments and microfluidic chip processing equipment development and production. We are committed to providing reliable microfluidic solutions to our global customers through technological innovation and high quality products to help them realize more accurate, reliable and efficient experiments and applications in the fields of biology, medicine, life sciences and environmental monitoring.
© 2025. All Rights Reserved. 苏ICP备2022036544号-1