1. Small molecule assays for single cells
Detection of small molecules in single-cell analysis is a necessary strategy to better understand organisms, explore cellular heterogeneity, and diagnose diseases early.
Intercellular small molecules such as lactate and reactive oxygen species are widely involved in cellular signaling pathways and play important roles in many physiological and pathological processes.
The research group developed a surface-enhanced Raman scattering (SERS)-microfluidic droplet platform that enables label-free simultaneous analysis of multiple metabolites at the single-cell level through a versatile magnetic SERS substrate consisting of Fe3O4 magnetic microspheres decorated with silver nanoparticles.
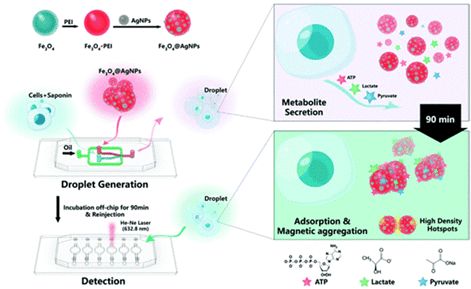
They achieved label-free, nondestructive, simultaneous determination of three single-cell metabolites: pyruvate, adenosine triphosphate, and lactate.
Their metal-magnetic composite substrate facilitates efficient adsorption of single-cell metabolites and rapid separation from complex matrices with high SERS sensitivity, making the SERS microdroplet platform a powerful tool for exploring single-cell heterogeneity at the metabolic level.
Single-cell droplets can also be categorized based on small molecule kinetics.
Ascorbic acid (AA), also known as vitamin C, is a strong antioxidant and a cofactor in enzyme reactions that protect against free radical-induced diseases, especially Parkinson's disease and cancer.
AA is effective in scavenging toxic free radicals and other reactive oxygen species associated with a wide range of tissue injuries and diseases, making it useful to develop simple, rapid, and easy-to-use analytical methods to measure AA highly selectively and reliably.
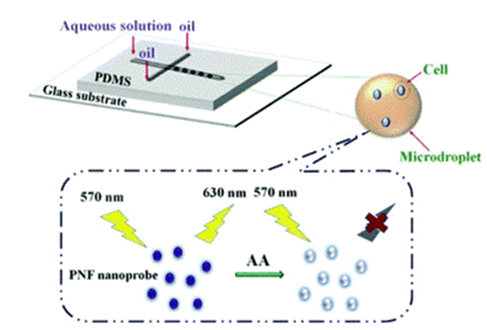
A related study reports a droplet microfluidic method for intracellular imaging of AA in living cells. By separating small microdroplets, they were able to rapidly detect and image AA in single cells.
2. Single-cell protein analysis
Proteins are biomolecules with specific functions in living cells that are directly related to the specificity of cell behavior and metabolism.
The proteins expressed in different cells are distinctly heterogeneous, with a wide variety of proteins within a single cell. Despite the abundance of protein types, the number of proteins with important functions is small.
Therefore, quantitative analysis of specific proteins based on single cells is important to reveal the heterogeneity of cells in a population and to improve the early diagnosis of diseases (e.g., cancer) caused by single cells or small cell populations.
Flow cytometry is a classical single-cell protein analysis method that allows the detection of specific functional proteins by immunolabeling multiple functional proteins.
Dynamic detection and analysis of specific proteins secreted by single cells based on conventional flow cytometry remains challenging due to the low abundance of secreted proteins, insufficient sensitivity, and the need for immobilization of cells.
Microfluidic droplet technology, with its small size, high detection sensitivity and high throughput, has a promising application in single-cell protein analysis.
Cytokines secreted by mammalian cells (e.g., immune cells, endothelial cells, and cancer cells) play a key role in infection, immune response, inflammation, and disease development.
Increased cytokine expression is associated with tumor vascularization and growth, which in turn generates new tumor blood vessels and enlarges existing ones.
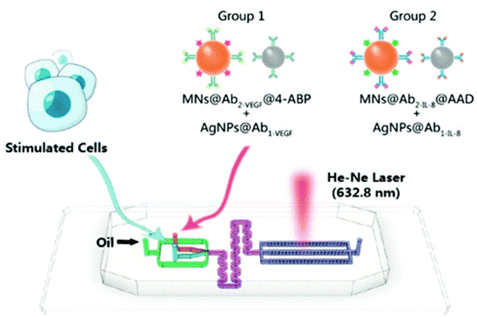
Panelists built a SERS droplet microfluidic platform for the rapid, ultrasensitive simultaneous detection of cytokines secreted by individual cancer cells, including vascular endothelial growth factor (VEGF) and IL-8.
Their findings suggest that cell-to-cell interactions can promote cancer cell angiogenesis by upregulating vascular endothelial growth factor and IL-8.
3. Single-cell drug screening analysis
Due to intercellular heterogeneity, different cells respond differently to certain drugs.
Although drug screening using conventional miniaturized microtiter plates is well established and simple to perform, drug screening for single cells still faces a number of technical hurdles, including uncontrolled evaporation of distributed liquids in an open environment.
Conventional miniaturized microplates can only statically culture cells, which limits their applicability because this method cannot mimic the natural extracellular microenvironment.
In contrast, microfluidic devices can be used for three-dimensional cell culture by continuous injection, which not only simulates the microenvironment associated with in vivo cell physiology, but also has the advantages of low sample consumption, low cost, and high throughput.
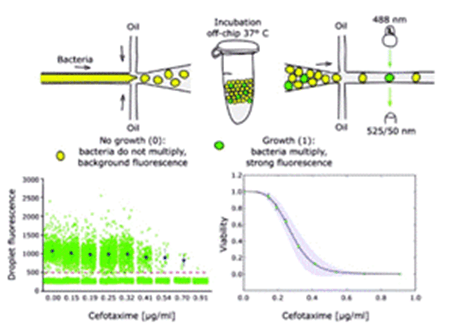
The investigators developed a droplet-based numerical minimum inhibitory concentration screen as a practical analytical platform to quantify the single-cell distribution of antibiotic phenotypic responses and to measure inoculum effects with high precision.
Based on the results obtained by treating Escherichia coli carrying β-lactamase with ceftazidime, they found that the efficacy of the antibiotic was determined by the amount of antibiotic per bacterial colony-forming unit rather than by the absolute concentration of antibiotic.
Unlike flow cytometry analysis, single-cell drug screening tests are performed using a microdroplet approach, in which selected compounds are added to isolated individual cells rather than to a population of cells.
This avoids cell-to-cell interactions, and because different cells have different sensitivities to drugs, drug-resistant cells can be isolated from cell populations for further analysis, potentially leading to the development of new drugs for the early treatment of cancer.
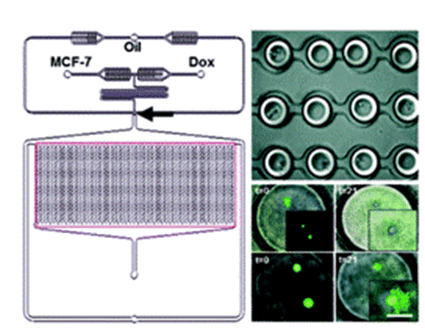
Researchers analyzed the uptake and cytotoxicity of doxorubicin in wild-type and doxorubicin-resistant breast cancer cells using an integrated microfluidic droplet array platform.
They found that drug-sensitive cells were more likely to die than drug-resistant cells in the presence or absence of doxorubicin (Dox).
4. Single-cell genetic analysis
Genetic differences between cells are of considerable importance in development, differentiation, signaling, and disease, making the development of single-cell genetic analysis methods important for exploring these life processes.
In addition to isolating single cells, droplet microfluidics has become an ideal tool for isolating single-cell genomes, making it an ideal technology for high-throughput studies of single-cell genomics in cell biology and clinical medicine.
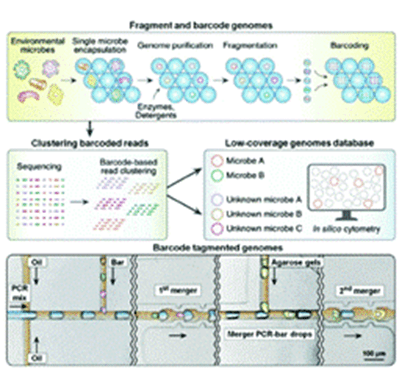
An experiment describes a method for ultra-high-throughput single-cell genome sequencing using microfluidic droplet technology.
In this method, individual cells are encapsulated in a microgel, which is permeable to molecules such as enzymes, detergents, and small molecules with hydrodynamic diameters smaller than their pore sizes, but also spatially traps large molecules such as genomic DNA.
The use of microgels facilitates the washing of encapsulated cells to perform the necessary steps such as cell lysis and genome processing, while maintaining the isolation of individual genomes.
Using this method, a series of operations such as cell lysis and genome processing can be easily performed on encapsulated cells while maintaining the separation between individual genomes.
By combining microgel and microfluidic droplet technologies, this method can process 50 000 cells in a short period of time, enabling ultra-high-throughput single-cell genome sequencing.
Site Search
Recommendations
© 2025. All Rights Reserved. 苏ICP备2022036544号-1













