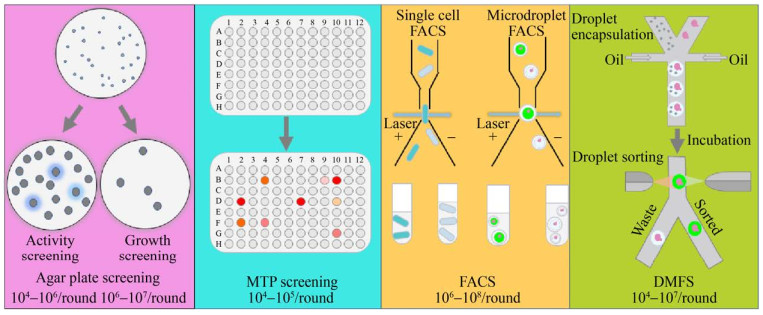
The traditional agar plate screening method and the microtiter plate screening method are the two commonly used screening methods today.
Agar plate screening for detection of activity by clear or color circles or for growth selection based on nutrient-deficient phenotypes or resistance is suitable for initial screening to exclude large numbers of inactive and low-activity mutants.
However, the agar plate screening method is not applicable to all modification targets and is difficult to accurately quantify.
For this reason, microtiter plate (MTP) screening methods based on the precise detection of the target product by fluorescence or absorbance have been developed, but their throughput is low and their operation is time-consuming.
To address these issues, ultra-high-throughput screening methods such as fluorescence-activated cell sorting (FACS) and droplet microfluidic sorting (DMFS) have been developed in recent years.
Agar plate screening
The agar plate method is a simple and direct screening method that has been widely used in the primary screening of mutant libraries for a variety of hydrolytic enzymes (e.g., lipases, esterases, proteases) and oxidoreductases (e.g., laccases).
The method is divided into two categories: phenotypic activity screening and phenotypic growth selection.
Phenotypic activity screening detects enzyme activity or target products by observing hydrolysis rings, color rings, or fluorescent products around colonies.
Phenotypic growth selection, on the other hand, is based on cell resistance to antibiotics or other deleterious substances, or through nutritionally deficient phenotypic complementation, based on growth in a selection medium.
However, the agar plate method is weak in visualizing differences between mutants and is only suitable for initial screening of mutant libraries. Mutants after screening still need to be accurately quantified by other assays such as the microtiter plate method.
The application of a digital image spectrophotometer has improved the sensitivity of the agar plate method, allowing a throughput of 10^5 clones per day to be screened.
If an agar plate screening method can be established based on the characteristics of the target enzyme or metabolite, there is no need to rely on complex instrumentation for ultra-high-throughput screening.
Microplate Screening
The microplate screening method is quantitatively analyzed by detecting the absorbance or fluorescence changes caused by the substrate or target product in the microplate, which ensures the accuracy and sensitivity of the screening, and it is the most commonly used screening method nowadays. The reaction volume is usually in the range of 100-200 μL, which saves the cost of reagents.
Commonly used microtiter plates are 96-well plates, compatible with automated monoclonal collection systems and automated liquid handling systems, with screening throughputs of up to 10^4 clones per day.
However, the introduction of mechanical automation equipment has increased the cost of experiments. To improve screening throughput and save time, 384-, 1536-, and 3456-well microtiter plates have also been used for the screening of target metabolites and enzymes, while saving the use of expensive substrates.
Nevertheless, the throughput of microtiter plate screening is still limited, which is not conducive to rapid screening of large-capacity mutation libraries.
Fluorescence Activated Cell Sorting (FACS)
FACS is a highly efficient single-cell sorting technique capable of sorting cells at rates up to 10^7 clones per hour based on cell size or fluorescence signal.
FACS also allows direct assignment of screened dominant mutants to microtiter plates for recovery and characterization.
Before performing FACS screening, it is important to establish the coupling of the enzyme activity phenotype to its encoding gene, translate the enzyme activity into a detectable fluorescent signal, and construct a physical link to the cell where the enzyme is located to ensure phenotypic and genotypic consistency.
According to the different forms of coupling of fluorescent products to enzymes and their encoding genes, FACS enzyme activity screening systems can be categorized into cell membrane surface display, intracellular fluorescent product enrichment, and fluorescent protein expression activity reporting.
For the detection of extracellular secretory enzymes or metabolites, cells can be embedded in water/oil/water bi-droplets or hydrogels to ensure genotypic and phenotypic correlation. The droplet-embedding technology broadens the application of FACS.
Droplet Microfluidic Screening (DMFS)
The DMFS (Droplet Microfluidic Sorting) method enables genotype-phenotype coupling by embedding individual cells in droplets at high frequency (>10 kHz) on a chip, and quantitative analysis and sorting by detecting material signals within the droplets, with a screening throughput of up to 10^5 clones per hour.
Water-in-oil droplets provide a nano-escalation to a pico-escalation reaction chamber, making this method suitable for screening not only intracellular enzymes or metabolites, but also extracellular secreted enzymes or metabolites.
Compared to conventional microliter to milliliter scale reaction systems, DMFS reduces the reaction volume by more than a million-fold, which greatly reduces the amount of reagents required and provides a significant advantage when using expensive substrates or reagents.
In addition, injection of analytical reagents, droplet fusion, and splitting can still be performed after monolayer droplet embedding, improving flexibility.
Water/oil/water double droplets formed by oil-in-water single droplets followed by aqueous phase embedding can also be screened by FACS, with a further increase in throughput to 10^7 clones per hour.
Hydrogel microdroplets formed using Microfluidic Chip can also be screened by FACS, and the hydrogel microdroplets are highly stable and suitable for prolonged cell culture.
However, the subsequent operations of double droplets and hydrogels after droplet formation (e.g., injection of analytical reagents, droplet fusion, splitting, etc.) are more difficult, which is not conducive to the flexible manipulation of the whole screening process.
Combining precision droplet manipulation with a rapid sorting system, DMFS has become a powerful tool for targeted modification of extracellular enzymes and metabolite mutant library screening.
Despite the significant advantages of DMFS in high-throughput screening of extracellular products, the process is complex and technical, making it difficult for beginners to operate, and the parameters of the DMFS screening system are highly variable for different target products or enzymes, requiring extensive system optimization.
About Us
DingXu (Suzhou) Microfluidics Technology Co., Ltd. is a high-tech enterprise dedicated to the field of microfluidics. We are committed to providing customers with comprehensive microfluidic solutions, including customized microfluidic chip development, surface modification, microfluidic chip processing equipment, and microfluidic instruments. Our team boasts extensive experience and technical expertise, continuously combining professional knowledge with innovative thinking to deliver high-quality solutions. We consistently prioritize customer-centric values, embrace self-challenges, and pursue excellence. Through professionalism, innovation, and collaboration, we aim to create greater value for our customers and contribute to a brighter future in the field of microfluidics.
© 2025. All Rights Reserved. 苏ICP备2022036544号-1