Many in vitro methods have long been used for high-throughput drug screening or toxicology testing. However, most of the currently available systems are only partial approximations of human biology and therefore have limited predictive power.
Indeed, these systems are either based on human cell cultures, which are unable to capture the complexity of cellular behavior in a three-dimensional (3D) environment, or on animal tissue fragments, which are three-dimensional in nature but only partially biologically similar to human tissues, and are unable to account for interactions with other organs.
To overcome these limitations, a new generation of bioreactors is being developed to generate multiple human cell-based tissue analogs in the same fluidic system to better reproduce the complexity and interconnectedness of human physiology.
These efforts aim to create multi-tissue organ systems (cardiovascular, gastrointestinal, musculoskeletal, etc.) and ultimately connect them into an interconnected human-on-a-chip device that realistically recreates the complexity of the human body's response to disease and potentially to drug therapy.
In this work, we focused on a specific bioreactor designed to generate multiple inputs/outputs of osteochondral structures, i.e., biphasic structures in which one side is essentially cartilage and the other is bone.
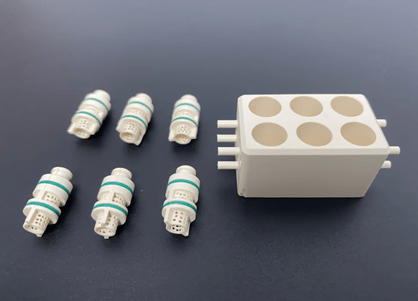
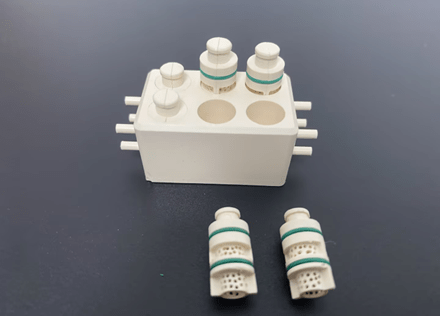
The case shown in Figure 1 was chosen for this Cell Culture Chip/Organoid Chip because it contains a two-chamber system to host a single biphasic tissue construct with different fluidics and a set of interconnected chambers with common fluids.
Starting with this particular bioreactor, we have developed a generalized method to simulate that multi-compartmental, interconnected microfluidics can be used to study human systems, and this technique employs the use of PEEK materials to prepare microfluidic PEEK chips.
The analysis of microphysiological osteochondral bioreactors (cell culture microarrays, or organoid microarrays) is aimed at the study of osteoarthritis (OA), the main pathology of the joints, which affects more than 33% of joints in people over 65 years of age.
This disease is characterized by a progressive degeneration of cartilage affecting all tissues and joints throughout the body, beginning long before clinical symptoms appear and eventually requiring joint replacement surgery.
This painful and disabling pathology with its high morbidity requires an understanding of the causes and mechanisms of its development in order to find restorative drug therapies to prevent or even cure the disease. Regenerate damaged tissue and ultimately avoid surgery.
This novel strategy respects the use of histotechnology and cell culture microarrays to generate a large number of identical in vitro constructs that can replicate the pathogenesis of joint diseases for the identification of therapeutic targets and drug screening.
Improvements in this regard have led to the development of a representative model of the interactions between cartilage and other joint tissues, particularly the subchondral bone. In fact, there is growing evidence of in vivo exchange of nutrients, cellular factors, and hormones between people.
Bone and cartilage. The osteochondral (OC) unit is recognized as the first of the medium-high-throughput systems studied in this work, referred to here as the high-throughput organoid core (HTB), which reflects the dynamic cartilage/bone interactions in health and disease.
It hosts a biphasic construct in a single chamber with separate fluidics for its cartilage and bone components, effectively creating a dual chamber setup (Figure 1). By this means, cartilage and bone will remain in contact and be able to send signals to each other while each is exposed to its ideal culture medium.
In addition, HTB allows the generation and culture of a large number of identical OC constructs at sizes similar to natural tissue biopsies. It is important to note that the physiological functions of the examined tissues are primarily weight-bearing and strength.
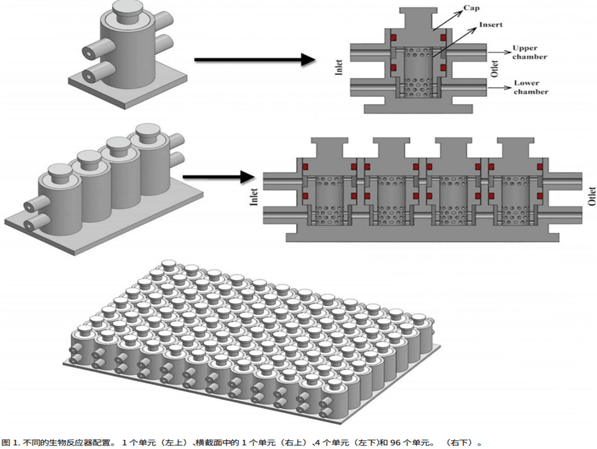
In order to achieve a high-throughput drug screening system, individual microfluidic PEEK chip dual chambers (bioreactor units) have been connected and assembled into a multi-unit system, which is sequentially and peacefully organized (Fig. 1).
In the 96-well design shown in Figure 1, the individual units are connected in series only, eight at a time, because this design is best suited for drug or toxicology screening; for example, to assess dose-response, each eight-unit array can accept different concentrations of the compound to be examined. Although possible, “parallel” connections have not yet been envisioned. The construction of each eight-unit array is indicated for repeated testing of multiple endpoints (e.g., histology, PCR, etc.).
The 3D CAD model of the microfluidic PEEK chip has the same configuration for each cell, is designed specifically for use in creating a combination of a soft gage and a gage, and consists of two inlets and two outlets consisting of cylindrical channels to ensure that fluid from the upstream cell is circulated downstream.
Each entry/exit channel has a length (L) of 5.3 mm and an inner diameter (d) of 1 mm. The perforated cylindrical inserter secures in place a footman that is 8.5 mm high and 3.75 mm wide. Each cell culture chip chamber is sealed by a gasket that seals the top lid and two o-rings (Figure 1).
This study will soon be extended to consider 8 rows of bioreactor cells. By aligning these lines with 12 parallel lines, a 96-unit plate can be obtained, which is a realistic prototype screen for a high-throughput organoid chip for drugs. The 3D structure is shown in the figure below.
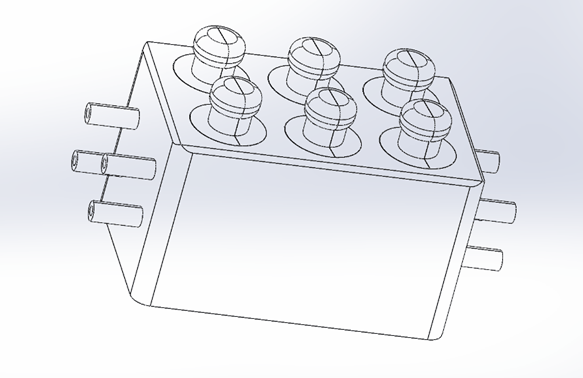
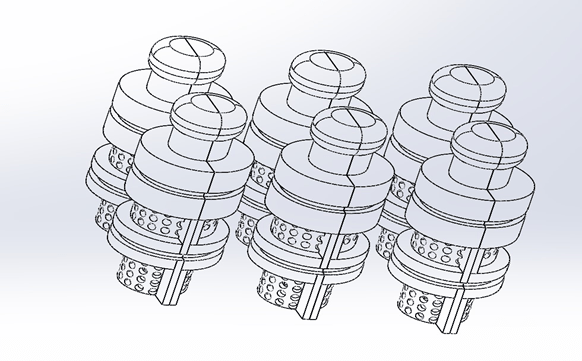
The microfluidic PEEK chip has a combination of an inlet, outlet, and external free-flow internal culture chamber (inset) with a porous media flow chamber. In each region, we assume that the flow is incompressible.
In order to achieve momentum balance, our approach employs a generalized scheme that incorporates the properties of both types of flows, and we will switch between them by appropriately tuning the problem parameters in each region.
This equation has the structure of the Brinkmann equation for flow in porous media, since it combines viscous terms, such as Stokes, and frictional terms, such as Darcy. In order to simulate the natural flow, the convective term plays an important role in the high Reynolds state is added also assuming static conditions.
Source of the above article:
1、 Iannetti L, D’Urso G, Conoscenti G, Cutrı`E, Tuan RS, Raimondi MT, et al. (2016) Distributed and Lumped Parameter Models for the Characterization of High Throughput Bioreactors.PLoS ONE 11(9): e0162774. doi:10.1371/journal.pone.0162774
(Article citation note: This article is for academic communication, if you need to delete please contact customer service)
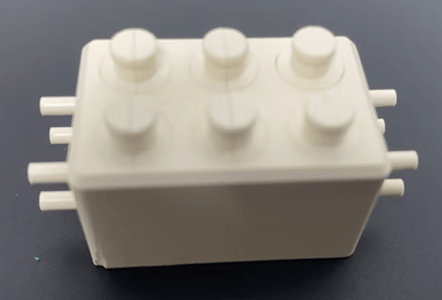
Microfluidic PEEK Chip/Cell Culture Chip/Organoid Chip Processing Physical Drawing

© 2025. All Rights Reserved. 苏ICP备2022036544号-1













